Mechanism of neurodegeneration-related protein aggregation

Protein misfolding and aggregation is a key event in the pathogenesis of neurodegenerative diseases such as Alzheimer (AD) and Parkinson diseases (PD) and widely considered as a suitable target for drug development against neurodegeneration. In our group, we employ solution-state NMR spectroscopy combined with other biophysical and computational techniques in order to obtain high-resolution mechanistic insights into protein aggregation. A particular focus of our research is to develop new NMR methods, e.g. high-pressure, singlet-state, fluorine and paramagnetic NMR, in order to expand our experimental access to protein motions underlying aggregation. The current system of interest is the AD-related amyloid-beta peptide, on which we apply the tailored NMR methods to investigate how its structure, dynamics, stability and aggregation are altered by AD-related mutations, modifications (such as phosphorylation) and metal-, peptide- and small-molecule binding.
For more details, please see our publications.
Biomolecular phase separation

Spatial arrangement of cellular components and activities is frequently achieved through a process known as liquid-liquid phase separation (LLPS). The membrane-less bodies formed through LLPS provide a microenvironment suitable for the activity of their biomolecular components. In our group, we are interested in developing new NMR methods to characterize biomolecular phase separation, especially in terms of the internal fluidity and component exchange of biomolecular condensates as well as mechanisms underlying their formation, maturation and dissolution. Of particular interest is to exploit the advantageous features of unconventional NMR nuclei, such as 19F and quadrupolar nuclei 17O and 23Na, and investigate how the physical properties of biomolecular condensates are affected by various biological regulatory factors.
For more details, please see our publications.
Protein dynamics in highly flexible protein systems
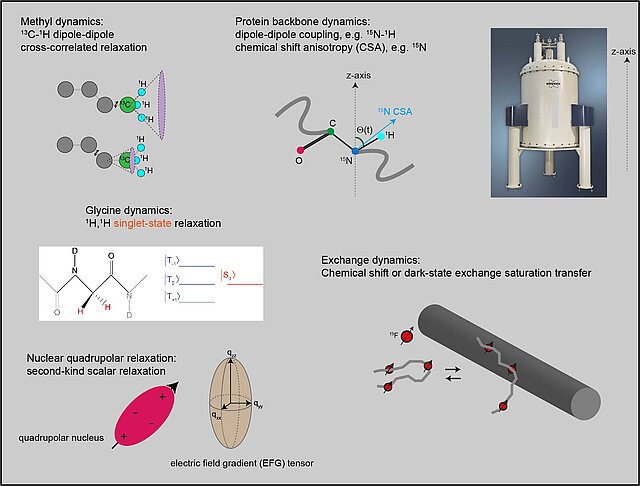
The function of macromolecular machines inside cells is often supported by the presence of intrinsically disordered proteins (IDP) regions, which their distance and reorientational dynamics govern the timescale and amplitude of the relative mobility of the constituent domains. Due to the lack of stable tertiary structures, the IDPs often populate a highly heterogeneous conformational ensemble and experience diverse range of motions at multiple length- and timescales. Our group is interested in developing a comprehensive conceptual framework and experimental approach to elucidate protein motions in IDPs, in the free form and in the context of larger systems. The main fronts of our research are: a) to overcome the current size- and timescale limitations of solution-state NMR methods through developing new NMR probes of protein dynamics, b) to evaluate our current understanding of the contribution of different relaxation mechanisms in IDPs, and c) to integrate the information obtained from NMR with the complementary information from other experimental and computational techniques.
For more details, please see our publications.

