Von der Sequenz zur Struktur und Funktion von nicht-kodierender RNA
Laut dem zentralen Dogma der Molekularbiologie agiert RNA nur als Boten-RNA (messenger RNA, mRNA) zwischen der genomischen DNA und der Protein-Biosynthese ("DNA macht RNA macht Protein"). Seit mehreren Jahren ist allerdings offensichtlich, dass RNA eine Vielzahl weiterer biologischer Aufgaben während Replikation, Transkription, Lokalisierung und Abbau biologischer Makromoleküle besitzt. Im Folgenden sind einige Beispiele aufgezählt:
- RNA kann enzymatische Funktion besitzen.
- Die Bildung der Peptidbindung im Ribosom wird durch die ribosomale RNA katalysiert.
- Spaltung und Ligation von RNA kann durch RNA katalysiert werden.
- RNA kann strukturelle Rollen besitzen.
- Die 7SL-RNA bildet mit mehreren Proteinen das Signal-Erkennungspartikel (signal recognition particle, SRP), das den Ort der intrazellulären Protein-Biosynthese beeinflusst.
- RNA kann die Gen-Expression beinflussen.
- MicroRNAs (miRNA), die in vielen Organismen genomisch kodiert sind, können die Translation von mRNA kontrollieren.
- "Silencing" RNA (siRNA) kann den Abbau von Fremd-RNA initialisieren.
- Ein Riboschalter (riboswitch) kann als Element einer mRNA deren Synthese oder Funktion zum Beispiel in Abhängigkeit von der Konzentration kleiner Moleküle oder auch der Temperatur (als RNA-Thermometer) regulieren.
Diese Klasse von RNAs wird als nicht-kodierende RNA (non-coding RNA, ncRNA) bezeichnet.
Für die biologische Funktion von nicht-kodierender RNA kann nicht nur deren Sequenz, sondern auch ihre zwei- oder dreidimensionale Struktur und ihre strukturelle Dynamik von entscheidender Bedeutung sein. In der Arbeitsgruppe entwickeln und testen wir bioinformatische Werkzeuge, die zum Beispiel für die Suche nach ncRNA-kodierenden Regionen in einem Genom oder für die Vorhersage von Struktur und Strukturdynamik benutzt werden können. Bespiele dafür sind im Folgenden gezeigt:

![]() StrAl
StrAl
ein Programm zum multiplen Alignieren von RNA-Sequenzen unter Berücksichtigung ihrer Basenpaarungswahrscheinlichkeiten.![]() Deniz, D., Wilm, A., Mainz, I. & Steger, G. (2006)
Deniz, D., Wilm, A., Mainz, I. & Steger, G. (2006)
STRAL: Progressive alignment of non-coding RNA using base pairing probability vectors in quadratic time.
Bioinformatics 22, 1593-1599.
![]() novoMIR
novoMIR
ein Programm zur de-novo-Vorhersage von prä-miRNA und miRNA/miRNA*-Bereichen in genomischer Pflanzen-DNA.![]() Teune, J.-H. & Steger, G. (2010)
Teune, J.-H. & Steger, G. (2010)
novoMIR: De Novo Prediction of MicroRNA-Coding Regions in a Single Plant-Genome.
J. Nucleic Acids 2010.

![]() ConStruct
ConStruct
ein Werkzeug zum interaktiven Alignieren und der Vorhersage von konservierter Sekundärstruktur.![]() Lück, R., Gräf, S. & Steger, G. (1999)
Lück, R., Gräf, S. & Steger, G. (1999)
ConStruct: a tool for thermodynamic controlled prediction of conserved secondary structure.
Nucleic Acids Research 27, 4208-4217.![]() Wilm, A., Linnenbrink, K. & Steger, G. (2008)
Wilm, A., Linnenbrink, K. & Steger, G. (2008)
ConStruct: improved construction of RNA consensus structures.
BMC Bioinformatics 9, 219.

![]() BRAliBase
BRAliBase
a "Benchchmark RNA Alignment database".![]() Wilm, A., Mainz, I. & Steger, G. (2006)
Wilm, A., Mainz, I. & Steger, G. (2006)
An enhanced RNA alignment benchmark for sequence alignment programs. Algorithms for Molecular Biology 1, 19.
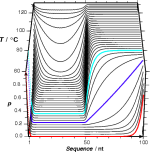
![]() poland
poland
ein Web-Service für die Berechnung der thermischen Stabilität doppelsträngiger Nukleinsäuren (DNA, dsRNA, RNA/DNA-Hybride).![]() Steger, G. (1994)
Steger, G. (1994)
Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction.
Nucleic Acids Research 22, 2760-2768.
Publikationen
- Bartl, J. and Zanini, M. and Bernardi, F. and Forget, A. and Blümel, L. and Talbot, J. and Picard D, Qin N, Cancila G, Gao Q, Nath S, Koumba IM, Wolter M, Kuonen F, Langini M, Beez, T, Munoz C, Pauck D, Marquardt V, Yu H, Souphron J, Korsch M, Mölders C, Berger D, Göbbels S, Meyer FD, Scheffler B, Rotblat B, Diederichs S, Ramaswamy V, Suzuki H, Oro A, Stühler K, Stefanski A, Fischer U, Leprivier G, Willbold D, Steger G, Buell A, Kool M, Lichter P, Pfister SM, Northcott PA, Taylor MD, Borkhardt A, Reifenberger G, Ayrault O, Remke M
The HHIP-AS1 lncRNA promotes tumorigenicity through stabilization of dynein complex 1 in human SHH-driven tumors.
Nat. Commun . 13, 4061 (2022)
https://doi.org/10.1038/s41467-022-31574-z
- Steger G
Predicting the Structure of a Viroid: Structure, Structure Distribution, Consensus Structure, and Structure Drawing
Methods in Molecular Biology 2316, 331-371 (2022)
https://doi.org/10.1007/978-1-0716-1464-8_26
- Steger G, Victor J
Design of a DNAzyme
Methods in Molecular Biology 2439, 47-63 (2022)
https://doi.org/10.1007/978-1-0716-2047-2_4
- Rosenbach H, Borggräfe J, Victor J, Wuebben C, Schiemann O, Hoyer W, Steger G, Etzkorn M, Span I
Influence of monovalent metal ions on metal binding and catalytic activity of the 10–23 DNAzyme
Biological Chemistry 402, 99-111 (2021)
https://doi.org/10.1515/hsz-2020-0207
- Steger G
Predicting the Structure of a Viroid
Methods in Molecular Biology 2316, 331-371 (2021)
https://doi.org/10.1007/978-1-0716-1464-8_26
- Rosenbach H, Victor J, Borggräfe J, Biehl R, Steger G, Etzkorn M, Span I.
Expanding crystallization tools for nucleic acid complexes using U1A protein variants
Journal of Structural Biology 210, 107480 (2020)
https://doi.org/10.1016/j.jsb.2020.107480
- Rosenbach H, Victor J, Etzkorn M, Steger G, Riesner D, Span I
Molecular Features and Metal Ions that Influence 10-23 DNAzyme Activity
Molecules 25, 3100 (2020)
https://doi.org/10.3390/molecules25133100
Special Issue "Advances in Catalytic DNA"
- Brass JRJ, Owens RA, Matoušek J, Steger G
Viroid quasispecies revealed by deep sequencing
RNA Biology 14, 317-325 (2017)
http://dx.doi.org/10.1080/15476286.2016.1272745
- Steger G
Modelling the three-dimensional structure of the right-terminal domain of pospiviroids.
Sci. Rep. 7, 711 (2017)
http://dx.doi.org/10.1038/s41598-017-00764-x
- Victor J, Steger G, Riesner D
Inability of DNAzymes to cleave RNA in vivo is due to limited Mg2+ concentration in cells
Eur Biophys J 47, 333-343 (2017)
https://doi.org/10.1007/s00249-017-1270-2
- Patra D, Fasold M, Langenberger D, Steger G, Grosse I, Stadler, PF
plantDARIO: web based quantitative and qualitative analysis of small RNA-seq data in plants
Frontiers in Plant Science 5, (2014)
http://dx.doi.org/10.3389/fpls.2014.00708
- Riesner D, Steger G
Temperature-gradient gel-electrophoresis.
In Bindereif, A., Hartmann, R., Schön, A. & Westhof, E. (eds), Handbook of RNA Biochemistry, Part II, 2nd edition, Wiley-VCH , 427-444 (2014)
http://dx.doi.org/10.1002/9783527647064.ch21
- Steger G
Secondary Structure Prediction.
In Bindereif, A., Hartmann, R., Schön, A. & Westhof, E. (eds), Handbook of RNA Biochemistry, Part III, 2nd edition, Wiley-VCH , 547-578 (2014)
http://dx.doi.org/10.1002/9783527647064.ch26
- Reinauer C, Censarek P, Kaber G, Weber AA, Steger G, Klamp T, Schrör K
Expression and translation of the COX-1b gene in human cells--no evidence of generation of COX-1b protein
Biol Chem 394, 753-760 (2013)
http://dx.doi.org/10.1515/hsz-2012-0309
- Steger G, Giegerich R
RNA structure prediction
in "RNA Structure and Folding: Biophysical Techniques and Prediction Methods" (Klostermeier D, Hammann C) de Gruyter, 335-362 (2013)
http://dx.doi.org/10.1515/9783110284959.335
- Aigner K, Dreßen F, Steger G
Methods for Predicting RNA Secondary Structure
In "Nucleic Acids and Molecular Biology: RNA 3D Structure Analysis and Prediction" (Leontis N, Westhof E) 27, 19-42 (2012)
http://dx.doi.org/10.1007/978-3-642-25740-7_3
- Janssen S, Schudoma C, Steger G, Giegerich R
Lost in folding space? Comparing four variants of the thermodynamic model for RNA secondary structure prediction.
BMC Bioinformatics 12, 429 (2011)
http://dx.doi.org/10.1186/1471-2105-12-429
- Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C
From alpaca to zebrafish: hammerhead ribozymes wherever you look
RNA 17, 21-26 (2011)
http://dx.doi.org/10.1261/rna.2429911
- Diermann N, Matousek J, Junge M, Riesner D, Steger G
Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato
Biol Chem 391, 1379-1390 (2010)
http://dx.doi.org/10.1515/BC.2010.148
- Teune, J-H, Steger G
novoMIR: de novo prediction of microRNA-coding regions in a single plant genome.
J. Nucleic Acids 2010, (2010)
http://dx.doi.org/10.4061/2010/495904
- Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, Westhof E
The RNA structure alignment ontology.
RNA 15, 1623-1631 (2009)
http://dx.doi.org/10.1261/rna.1601409
- Fu Y, Bannach O, Chen H, Teune JH, Schmitz A, Steger G, Xiong L, Barbazuk WB
Alternative Splicing of Anciently Exonized 5S rRNA Regulates Plant Transcription Factor TFIIIA.
Genome Research 19, 913-21 (2009)
http://dx.doi.org/10.1101/gr.086876.108
- Wilm A, Linnenbrink K, Steger G
ConStruct: improved construction of RNA consensus structures
BMC Bioinformatics 9, 219 (2008)
http://dx.doi.org/10.1186/1471-2105-9-219
- Censarek P, Steger G, Paolini C, Hohlfeld T, Grosser T, Zimmermann N, Fleckenstein D, Schrör K, Weber A-A
Alternative splicing of platelet cyclooxygenase-2 mRNA in patients after coronary artery bypass grafting
Thrombosis and Haemostasis 98, 1309-1315 (2007)
http://dx.doi.org/10.1160/TH07-05-0346
- Schmitz M, Steger G
Potato spindle tuber viroid (PSTVd).
Plant viruses. 1, 106-115 (2007)
- Dalli D, Wilm A, Mainz I, Steger, G
STRAL: Progressive alignment of non-coding RNA using base pairing probability vectors in quadratic time.
Bioinformatics 22, 1593-1599 (2006)
https://doi.org/10.1093/bioinformatics/btl142
- Gräf S, Teune J-H, Strothmann D, Kurtz S, Steger, G
A computational approach to search for non-coding RNAs in large genomic data.
In Nellen, W. & Hammann, C. (Hrsg.), Small RNAs: Analysis and Regulatory functions., Nucleic Acids and Molecular Biology Series, Band 17. Springer Verlag , 74-75 (2006)
- Marszalkowski M, Teune J-H, Steger G, Hartmann RK, Willkomm, DK
Thermostable RNase P RNAs lacking P18 identified in the Aquificales.
RNA 12, 1915-1921 (2006)
http://dx.doi.org/10.1261/rna.242806
- Wilm A, Mainz I, Steger G
An enhanced RNA alignment benchmark for sequence alignment programs.
Algorithms Mol Biol 1, 19 (2006)
http://dx.doi.org/10.1186/1748-7188-1-19
- Gardner PP, Wilm A, Washietl, S
A benchmark of multiple sequence alignment programs upon structural RNAs.
Nucleic Acids Res. 33, 2433-2439 (2005)
http://dx.doi.org/10.1093/nar/gki541
- Gräf S, Przybilski R, Steger G, Hammann C
A database search for hammerhead ribozyme motifs.
Biochem. Soc. Trans. 33, 477-478 (2005)
Publisher
- Przybilski R, Gräf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann, C
Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana.
Plant Cell 17, 1877-1885 (2005)
http://dx.doi.org/10.1105/tpc.105.032730
- Gräf S, Borisova BE, Nellen W, Steger G, Hammann, C
A database search for double-strand containing RNAs in Dictyostelium discoideum.
Biol. Chem. 385, 961-965 (2004)
http://dx.doi.org/10.1515/BC.2004.125
- Riesner D, Steger G
Temperature-gradient gel-electrophoresis.
In Bindereif, A., Hartmann, R., Schön, A. & Westhof, E. (Hrsg.), Handbook of RNA Biochemistry, Wiley-VCH , 398-414 (2004)
- Steger, G
Secondary Structure Prediction.
In Bindereif, A., Hartmann, R., Schön, A. & Westhof, E. (Hrsg.), Handbook of RNA Biochemistry, Wiley-VCH , 513-535 (2004)
- Dingley AJ, Steger G, Esters B, Riesner D, Grzesiek S
Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis.
J. Mol. Biol. 334, 751-767 (2003)
http://dx.doi.org/10.1016/j.jmb.2003.10.015
- Steger G
Bioinformatik: Methoden zur Vorhersage von RNA- und Proteinstruktur.
Birkhäuser Verlag Basel , (2003)
- Steger G, Riesner D
Properties of Viroids: Molecular characteristics.
In "Viroids" (Hadidi A, Flores R, Randles JW, Semancik JS, eds.) CSIRO Publishing , 15-29 (2003)
- Yukawa Y, Matousek J, Grimm M, Vrba L, Steger G, Sugiura M, Beier H
Plant 7SL RNA and tRNA(Tyr) genes with inserted antisense sequences are efficiently expressed in an in vitro system from Nicotiana tabacum cells.
Plant Mol. Biol. 50, 713-723 (2002)
http://dx.doi.org/10.1023/A:1019905730397
- Gräf S, Strothmann D, Kurtz S, Steger, G
HyPaLib: a database of RNAs and RNA structural elements defined by hybrid patterns.
Nucleic Acids Res. 29, 196-198 (2001)
http://dx.doi.org/10.1093/nar/29.1.196
- Antal M, Mougin A, Kis M, Boros E, Steger G, Jakab G, Solymosy F, Branlant C
Molecular characterization at the RNA and gene levels of U3 snoRNA from a unicellular green alga, Chlamydomonas reinhardtii
Nucleic Acids Res 28, 2959-2968 (2000)
http://dx.doi.org/10.1093/nar/28.15.2959
- Lück R, Gräf S, Steger G
ConStruct: a tool for thermodynamic controlled prediction of conserved secondary structure
Nucleic Acids Res 27, 4208-4217 (1999)
PubMed
- Matoušek J, Junker V, Vrba L, Schubert J, Patza, J, Steger G
Molecular characterization and genome organization of 7SL RNA genes from hop (Humulus lupulus L)
Gene 239, 173-183 (1999)
http://dx.doi.org/10.1016/S0378-1119(99)00352-2
- Repsilber D, Wiese U, Rachen M, Schröder AR, Riesner D, Steger G
Formation of metastable RNA structures by sequential folding during transcription: Time-resolved structural analysis of potato spindle tuber viroid (-)-stranded RNA by temperature-gradient gel electrophoresis.
RNA 5, 574-584 (1999)
Publisher
- Lück R, Steger G, Riesner D
Thermodynamic prediction of conserved secondary structure: Application to RRE-element of HIV, tRNA-like element of CMV, and mRNA of prion protein.
J Mol Biol 258, 813-826 (1996)
- Owens RA, Steger G, Hu Y, Fels A, Hammond RW, Riesner D
RNA structural features responsible for potato spindle tuber viroid pathogenicity.
Virology 222, 144-158 (1996)
http://dx.doi.org/10.1006/viro.1996.0405
- Schmitz M, Steger G
Description of RNA folding by ``simulated annealing''.
J Mol Biol 255, 254-266 (1996)
- Chen X, Baumstark T, Steger G, Riesner D
High resolution SSCP by optimization of the temperature by transverse TGGE
Nucleic Acids Res 23(21), 4524-5 (1995)
- Gruner R, Fels A, Qu F, Zimmat R, Steger G, Riesner D
Interdependence of pathogenicity and replicability with potato spindle tuber viroid
Virology 209(1), 60-9 (1995)
- Riesner D, Baumstark T, Qu F, Klahn T, Loss P, Rosenbaum V, Schmitz M, Steger G
Physical basis and biological examples of metastable RNA structures.
In "Structural tools for the analysis of protein-nucleic acids complexes. Advances in Life Sciences" (Lilley D, Heumann H, Suck D, eds.) Birkhäuser , 401-435 (1992)
- Schmitz M, Steger G
Base-pair probability profiles of RNA secondary structures.
CABIOS 8, 389-399 (1992)
- Steger G, Baumstark T, Mörchen M, Tabler M, Tsagris M, Sänger HL, Riesner D
Structural requirements for viroid processing by RNase T1.
J Mol Biol 227, 719-737 (1992)
- Loss P, Schmitz M, Steger G, Riesner D
Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA
EMBO J 10(3), 719-27 (1991)
- Owens RA, Thompson SM, Steger G
Effects of random mutagenesis upon potato spindle tuber viroid replication and symptom expression.
Virology 185, 18-31 (1991)
- Riesner D, Henco K, Steger G
Temperature-gradient gel electrophoresis: A method for the analysis of conformational transitions and mutations in nucleic acids and proteins.
In "Advances in electrophoresis" (Chrambach A, Dunn MJ, Radola BJ, eds.) VCH Weinheim Vol. 4, 169-250 (1991)
- Riesner D, Steger G
Viroids and viroid-like RNA.
In "Nucleic Acids, Subvol. d, Physical Data II, Theoretical Investigations, Landolt-Börnstein. Group VII Biophysics, 1, 194-243 (1990)
- Rubino L, Tousignant ME, Steger G, Kaper JM
Nucleotide sequence and structural analysis of two satellite RNAs associated with chicory yellow mottle virus (CYMV).
J Gen Virol 71, 1897-1904 (1990)
- Riesner D, Steger G, Zimmat R, Owens RA, Wagenhofer M, Hillen W, Vollbach S, Henco K
Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions
Electrophoresis 10(5-6), 377-89 (1989)
- Riesner D, Steger G, Zimmat R, Owens RA, Wagenhöfer M, Hillen W, Vollbach S, Henco K
Temperature-gradient gel electrophoresis: Analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions.
Electrophoresis 10, 377-389 (1989)
- Hecker R, Wang ZM, Steger G, Riesner D
Analysis of RNA structures by temperature-gradient gel electrophoresis: viroid replication and processing
Gene 72(1-2), 59-74 (1988)
- Kaper JM, Tousignant ME, Steger G
Nucleotide sequence predicts circularity and self-cleavage of 300-ribonucleotide satellite of arabis mosaic virus.
Biochem Biophys Res Commun 154, 318-325 (1988)
- Steger G, Tien P, Kaper J, Riesner D
Double-stranded cucumovirus associated RNA 5: Which sequence variations may be detected by optical melting and temperature-gradient gel electrophoresis?
Nucleic Acids Res 15, 5085-5103 (1987)
- Tien P, Steger G, Rosenbaum V, Kaper J, Riesner D
Double-stranded cucumovirus associated RNA 5: Experimental analysis of necrogenic and non-necrogenic variants by temperature-gradient gel electrophoresis.
Nucleic Acids Res 15, 5069-5083 (1987)
- Steger G, Tabler M, Brüggemann W, Colpan M, Klotz G, Sänger HL, Riesner D
Structure of viroid replicative intermediates: Physico-chemical studies on SP6 transcripts of cloned oligomeric potato spindle tuber viroid.
Nucleic Acids Res 14, 9613-9630 (1986)
- Werntges H, Steger G, Riesner D, Fritz HJ
Mismatches in DNA double strands: Thermodynamic parameters and their correlation to repair efficiencies.
Nucleic Acids Res 14, 3773-3790 (1986)
- Steger G, Hofmann H, Förtsch J, Gross HJ, Randles JW, Sänger HL, Riesner D
Conformational transitions in viroids and virusoids: Comparison of results from energy minimization algorithm and from experimental data.
J Biomol Struct Dyn 2, 543-571 (1984)
- Riesner D, Colpan M, Goodman TC, Nagel L, Schumacher J, Steger G, Hofmann H
Dynamics and interactions of viroids.
J Biomol Struct Dyn 1, 669-688 (1983)
- Riesner D, Steger G, Schumacher J, Gross HJ, Randles JW, Sänger HL
Structure and function of viroids.
Biophys Struct Mech 9, 145-170 (1983)
- Steger G, Müller H, Riesner D
Helix-coil transitions in double-stranded viral RNA: Fine resolution melting and ionic strength dependence.
Biochim Biophys Acta 606, 274-284 (1980)

